Active
Soil2Milk: Enhancing Soil to Milk Chain Sustainability
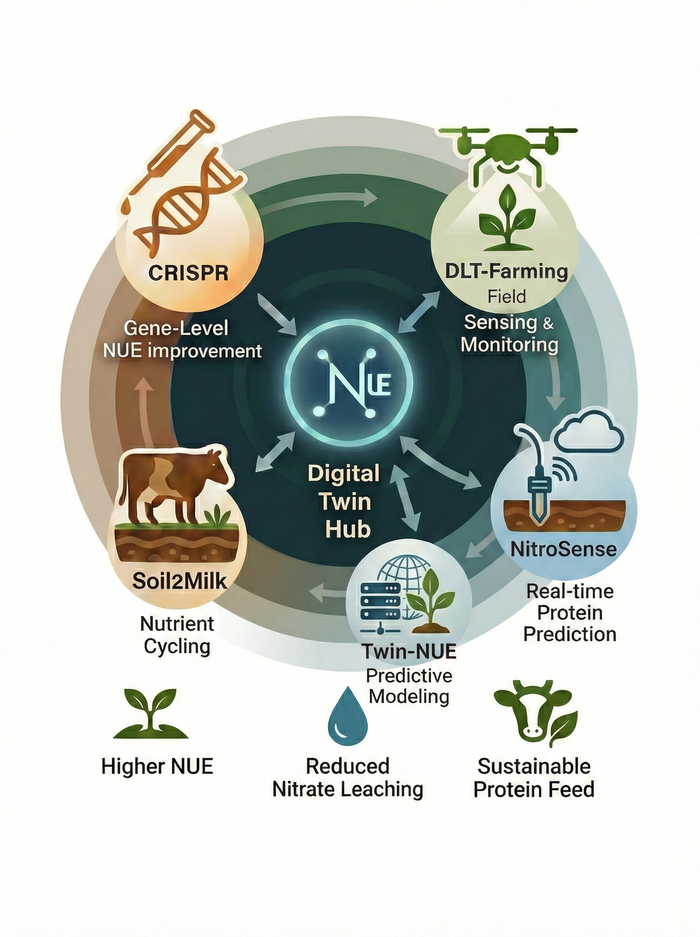
Norway’s agricultural sector faces significant environmental challenges from intensive nitrogen fertilization in forage production. Soil2Milk generates a sustainable feed value chain by enhancing nitrogen use efficiency (NUE) of forage crops and lowering enteric methane emissions. The project compares mixtures of perennial ryegrass/red clover and perennial ryegrass/birdsfoot trefoil (Lotus corniculatus) against traditional monocultures under two fertilizer regimes. Key objectives include investigating N₂O emissions and nitrogen fixation in grass–legume mixtures, identifying contrasting NUE varieties and applying CRISPR to the N biochemical pathway, assessing the impact of forage quality on dairy cow methane and milk yield, and modelling GHG emission intensities across the soil-to-milk value chain using the HolosNor model and machine learning. Partners include TINE, YARA, Graminor, NLR, EMBRAPA (Brazil), and multiple NMBU departments.

Soil2Milk kickoff group photo

Soil2Milk kickoff meeting
Active
ProteinSense: More Protein from Grass, Less Import of Concentrates
Protein levels in Norwegian forage grasses drop rapidly when harvest is delayed, increasing dependence on imported soy and rapeseed meal. ProteinSense addresses this by combining daily field sampling of perennial ryegrass varieties under three nitrogen regimes (20, 30, 33 kg N/daa) with drone-based multispectral/hyperspectral imaging, handheld NIRS, and laboratory protein analysis (Kjeldahl). The resulting dataset feeds AI models that predict crude protein content in real time and provide 3–5 day harvest forecasts, giving farmers a practical “harvest alert” to capture peak protein and reduce concentrate imports.
📅 2026–2028
💰 0.75 MNOK · FFL/JA (RCN) · Project Leader
Active
TWIN-NUE: TraitFinder-Enabled Digital Twins for Nitrogen Use Efficiency
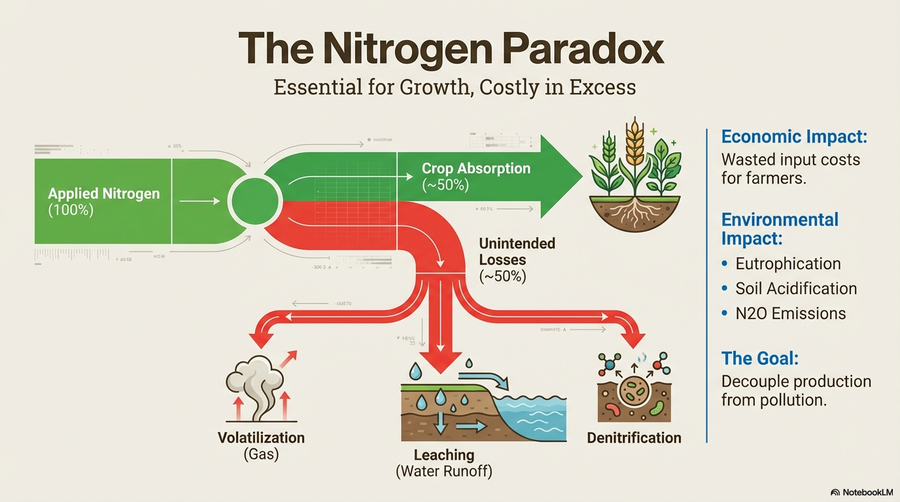
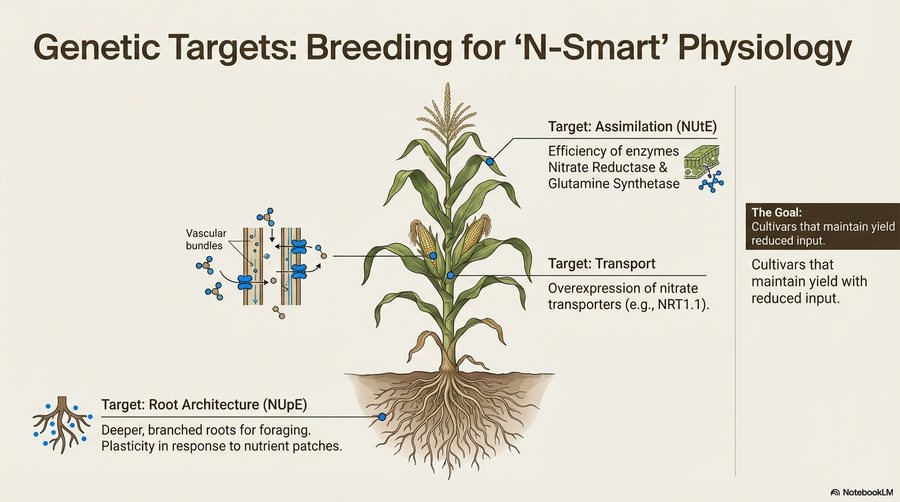
Only 30–50% of applied fertilizer nitrogen is recovered in crop biomass; the remainder contributes to nutrient leaching, N₂O emissions, and avoidable costs. TWIN-NUE exploits the Norwegian Plant Phenotyping Infrastructure (PheNo) and its TraitFinder system—a multispectral 3D laser scanner delivering daily, non-invasive measurements of canopy geometry, chlorophyll, and biomass. Controlled greenhouse trials with diverse genotypes of perennial ryegrass (Lolium perenne) and oat (Avena sativa) under low, standard, and high N regimes create high-resolution datasets. AI models build digital twins that simulate nitrogen scenarios to support genotype selection for optimal NUE, yield, and quality.
📅 2026–2029
💰 5.5 MNOK · PheNo / NMBU · Project Leader
Active
DLT-Farming: Data-Led Transformation for Sustainable Forage Grass Farming
Grassland-based forage production is critical for Norwegian agriculture, yet expansion of perennial ryegrass cultivation increases GHG emissions and nitrogen leaching. DLT-Farming creates a sustainable data-led transformation solution using robotics, energy-efficient IoT sensor networks, genomics, and AI/ML. Field trials with 40 cultivars from five countries under three fertilizer regimes at NMBU Ås and Graminor Hamar evaluate NUE through integrated phenomics and genomics. The project delivers an AI/ML data analytics platform for autonomous processing of drone and robot sensor data, a real-time reporting application for dry matter yield and forage quality, and GWAS-based identification of NUE genes to accelerate breeding. Partners: Accenture, BioDrone, and Graminor.
📅 2024–2028
💰 12 MNOK · FFL/JA (RCN) · Project Leader
Active
NitroGenEdit: Less Nitrogen, More Yield and High Quality Grass
Traditional breeding for high yield under low nitrogen input is slow and costly. NitroGenEdit develops innovative perennial ryegrass varieties that use less nitrogen while delivering higher yield and quality through CRISPR-mediated genome editing. The project characterizes Norwegian ryegrass cultivars for nitrogen uptake under varied nitrogen levels in hydroponic systems (WP-1), develops CRISPR constructs targeting key nitrogen transporter and metabolism genes (GS1, GS2, NiR, NRT1.1, NRT2B) in diploid and tetraploid cultivars (WP-2), and disseminates findings to farmers, breeders, and academia (WP-3). Nine CRISPR/Cas9 constructs have been designed and verified, with GS1 overexpression and NRT1.1 modulation identified as promising targets. The findings establish a foundation for lab-to-field scaling in larger future projects.
📅 2024–2026
💰 0.75 MNOK · FFL/JA · Project Leader
Active
GE-Sustain: Sustainable Potato Production by Precision Breeding
Applying precision breeding techniques for sustainable potato production, characterizing genes related to late blight resistance and industrial quality traits such as asparagine and glycoalkaloid content. Contributing as work package leader within a larger consortium funded by the Research Council of Norway.
📅 2024–2028
💰 3 MNOK (16 MNOK total) · FFL/JA (RCN) · WP Leader
 Field trials evaluating NUE genotypes at NMBU Ås
Field trials evaluating NUE genotypes at NMBU Ås
 IoT sensors capturing real-time soil nitrogen data
IoT sensors capturing real-time soil nitrogen data
 Dr. Kovi presenting the integrated NUE research vision
Dr. Kovi presenting the integrated NUE research vision

