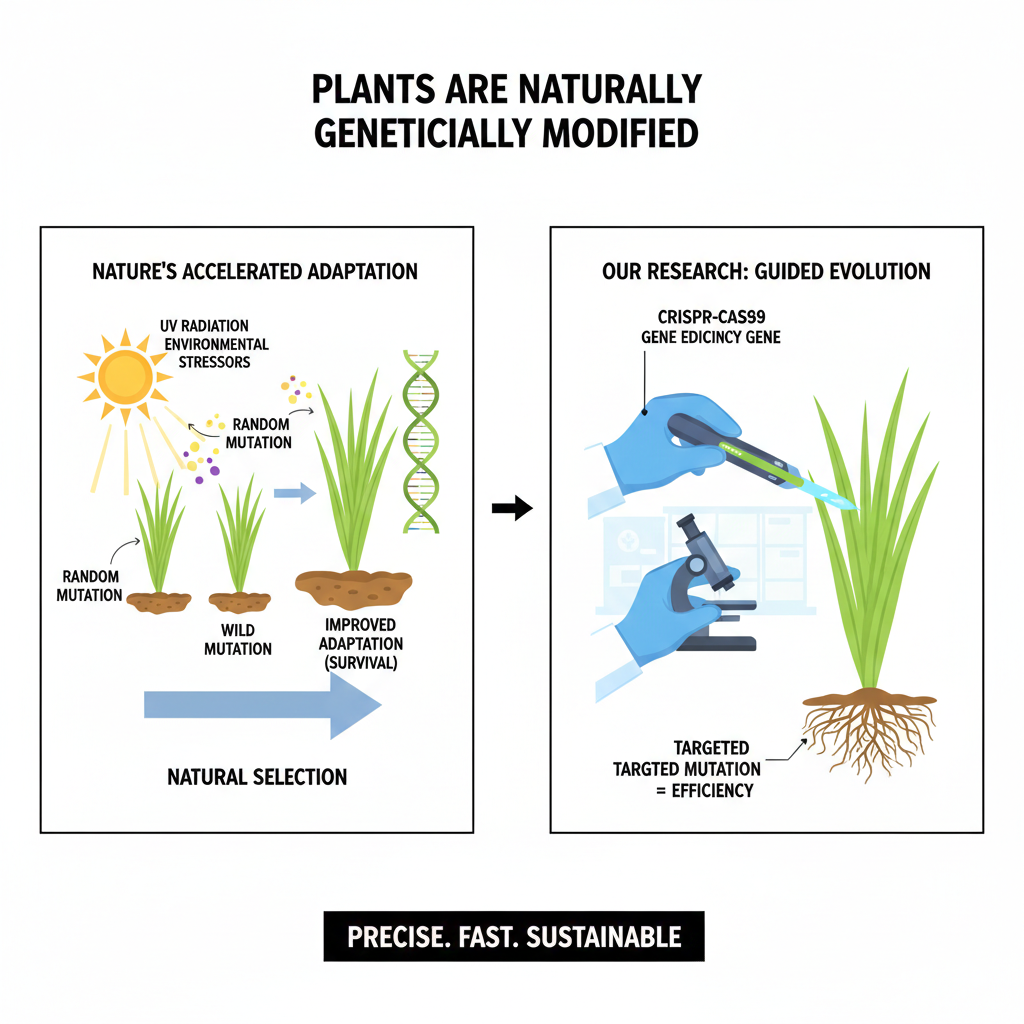
Functional Genomics: From Gene Discovery to Precision Breeding
Our functional genomics work aims to characterize gene function through RNA-seq transcriptomics, differential gene expression analysis, and CRISPR/Cas9 gene editing. We study key nitrogen metabolism genes (GS1, GS2, NiR, NRT1.1, NRT2B) and genes regulated in response to cold, drought, providing insights for targeted crop improvement through precision breeding.
RNA-seq experiments are carried out under controlled conditions with multiple nitrogen regimes and stress treatments, generating comprehensive transcriptomic profiles that reveal how plants reprogram gene expression in response to environmental challenges. Differential expression analysis identifies candidate genes whose activity is tightly linked to nitrogen uptake efficiency, cold acclimation pathways.
On the genome editing front, we have established successful transformation protocols for perennial ryegrass. Currently we designed nine CRISPR/Cas9 constructs targeting critical nitrogen transporter and assimilation genes in perennial ryegrass. GS1 overexpression and NRT1.1 modulation have emerged as particularly promising targets for enhancing nitrogen use efficiency without sacrificing yield. Once these mutants are derived, these will be characterized in hydroponic and greenhouse systems, laying the groundwork for future field-scale validation.
Together, our transcriptomic and gene editing approaches create a powerful feedback loop, identifying candidate genes through expression profiling and then functionally validating them through precise genomic modifications.