GWAS & Genomics Workflow
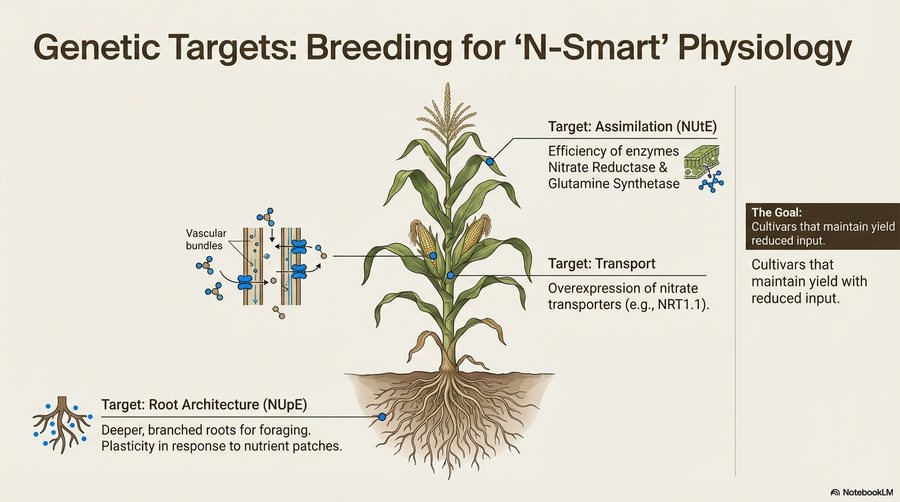
We investigate the genetic basis of critical agronomic traits to drive sustainable crop improvement. Using whole-genome sequencing, genotyping-by-sequencing (GBS), and genome-wide association studies (GWAS), we identify genomic variants linked to Nitrogen Use Efficiency (NUE), winter hardiness, yield, and quality. A primary focus of our lab is improving NUE in forage grasses to an optimal 70–80% range for sustainable agriculture.
Our genomics pipeline spans from DNA extraction and library preparation to advanced bioinformatic analysis and variant calling. We leverage reference genomes, resequencing, and transcriptomics (RNA-seq) for:
- Forage Grasses (Lolium perenne & Phleum pratense): Mapping quantitative trait loci (QTL) and utilizing RNA-seq to understand gene expression patterns related to nitrogen uptake, assimilation, and remobilization.
- Potato (Solanum tuberosum): Utilizing resequencing data from 10 diverse genotypes and RNA-seq to discover and characterize candidate genes underlying disease resistance and tuber quality.
By combining high-throughput genotyping and expression profiling with carefully designed field experiments, we dissect the genetic and regulatory control of complex traits. This integrated knowledge feeds directly into our genomic selection and functional genomics programs, creating a seamless pathway from gene discovery to practical breeding applications.